The eHealth Knowledge Discovery Task¶
The eHealth-KD task consists in automatically indentifying all the relevant semantic entities in a Spanish language document, as well as semantic relations among them.
To facilitate both development and evaluation, this task is subdivided into two subtasks:
-
Given a list of eHealth documents written in Spanish, the goal of this subtask is to identify all the entities per document and their types. These entities are all the relevant terms (single word or multiple words) that represent semantically important elements in a sentence. The following figure shows the relevant entities that appear in a set of example sentences.
-
Subtask B: Relation extraction
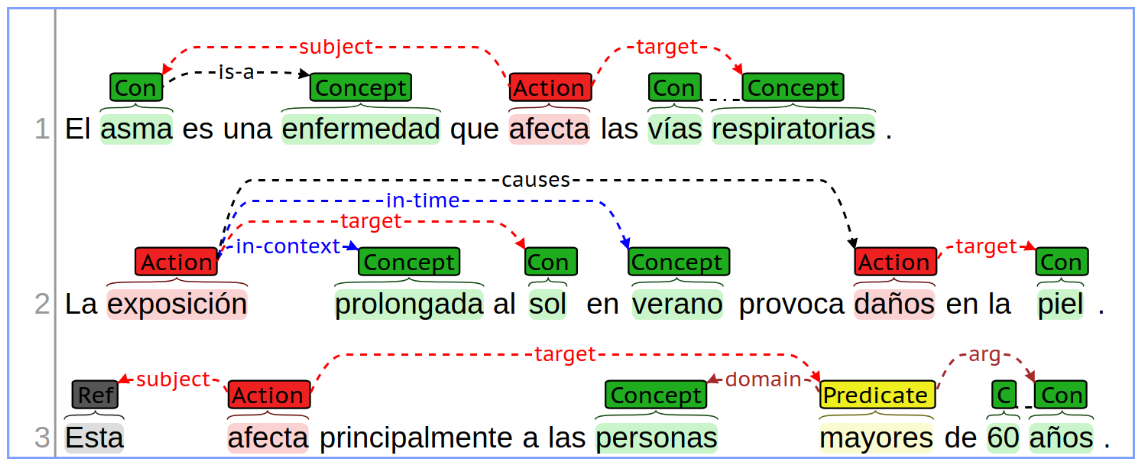
Subtask B continues from the output of Subtask A, by linking the entities detected and labelled in the input document. The purpose of this subtask is to recognize all relevant semantic relationships between the entities recognized. Eight of the thirteen semantic relations defined for this challenge can be identified in the following example:
Input and output format¶
Input files in the eHealth-KD 2020 are plain text files in UTF-8 format with one sentence per line. Sentences have not been preprocesed in any sense.
The output is always one file in BRAT standoff format, where each line represents either an entity or a relation. All details about the required format are available in the link above. We provide Python scripts to read and write this format in the repository of the eHealth-KD 2020 Challenge. More details in the Resources section.
An example output file for the annotation represented in the previous images is provided below:
The order in which the entities and relations appear in the output file is irrelevant. It is only important to be consistent with respect to identifiers. Each entity has a unique identifier that is used in relation annotations to reference it. Your output file can have different identifiers than the gold output, and the evaluation scripts will be able to correctly find matching annotations.
For example, if in your output the entity "asma" has ID T3 instead of T1 as in the previous example, and there appears a relation annotation R1 is-a Arg1:T3 Arg2:T2, this will be correctly matched with the corresponding gold annotation. Relation IDs are necesary for the BRAT format to be correctly parsed, but are irrelevant with respect to the evaluation of the task. Feel free to simply use autoincremental identifiers, or use the provided Python scripts, which already take care of all these details.
Important: Note about negated concepts and other attributes¶
The eHealth-KD corpus considers negated actions, which are manually annotated in the corresponding Brat files, as well as other attributes to indicate emphasis, uncertainty, etc. However, for evaluations purposes, we are not considering the annotation of negation or any other attribute.
This means that, in the corpus, you will find sentences with negated concepts, such as: "No existe un tratamiento que restablezca la función ovárica normal.". In this and similar sentences, we still expect that your system recognizes existe as Action and tratamiento as Target, as though if the negation did not exist.